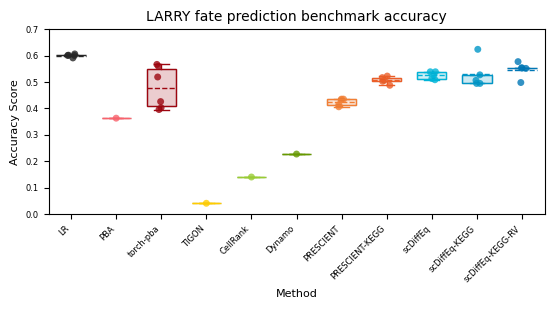
Figure 2d#
Aggregating fate prediction accuracy across methods
Import packages#
[8]:
import ABCParse
import anndata
import cellplots as cp
import glob
import larry
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import pathlib
import scdiffeq as sdq
import scdiffeq_analyses as sdq_an
import sklearn
from typing import List, Optional
Helper functions#
[55]:
def _read_process_F_hat(path, pickle=False, filter_nan=True):
if pickle:
F_hat = pd.read_pickle(path)[0]
else:
F_hat = pd.read_csv(path, index_col=0)
F_hat_filt = F_hat.copy().drop("Undifferentiated", axis=1)
F_hat_filt_norm = F_hat_filt.div(F_hat_filt.sum(1), axis=0).fillna(0)
if filter_nan:
F_hat_filt_norm = replace_undiff(F_hat_filt_norm)
F_hat_filt_norm.index = F_obs.index # F_hat_filt_norm.index.astype(str)
return F_hat_filt_norm
def replace_undiff(F_hat_filt_norm):
# create a temp vector to add undiff label
undiff = np.zeros(len(F_hat_filt_norm))
replace_idx = np.where(F_hat_filt_norm.sum(1) == 0)
# print(f"{replace_idx[0].shape[0]} non-fates")
undiff[replace_idx] = 1
# add to normalized matrix
F_hat_filt_norm["Undifferentiated"] = undiff
return F_hat_filt_norm
def compute_accuracy(F_obs, F_hat):
y_true, y_pred = F_obs.idxmax(1).tolist(), F_hat.idxmax(1).tolist()
return sklearn.metrics.accuracy_score(y_true, y_pred)
def get_organized_results(project_path):
paths = list(project_path.glob("*/fate_prediction_metrics/*/F_hat.unfiltered.csv"))
OrgResults = {}
for path in paths:
path_ = pathlib.Path(path)
version = path_.parent.parent.parent.name
if not version in OrgResults:
OrgResults[version] = {}
ckpt_name = path_.parent.name
F_hat = _read_process_F_hat(path_, pickle=False)
F_hat.index = F_hat.index.astype(str)
acc = larry.tasks.fate_prediction.metrics.multi_idx_accuracy(F_obs, F_hat)
train_acc = acc.loc["unique_train.all_fates"].iloc[0]
test_acc = acc.loc["unique_test.all_fates"].iloc[0]
OrgResults[version][ckpt_name] = {"train": train_acc, "test": test_acc}
acc_ = compute_accuracy(F_obs, F_hat)
# print("| Accuracy: Train: {:.5f}, Test: {:.3f} |".format(train_acc, test_acc), version, ckpt_name)
return OrgResults
def get_best_results(OrgResults):
BestResults = {}
for key, val in OrgResults.items():
version_accuracy = pd.DataFrame(val).T
best_ckpt = version_accuracy["train"].idxmax()
best_test = version_accuracy.loc[best_ckpt]["test"]
# print(key, best_ckpt, best_test)
# best_.append()
BestResults[key] = best_test
return BestResults
Load adata#
[72]:
adata = anndata.read_h5ad("/home/mvinyard/data/adata.LARRY_train.19MARCH2024.h5ad")
F_obs (pd.DataFrame)#
[62]:
F_obs = larry.tasks.fate_prediction.F_obs
Linear regression (LR)#
[11]:
LR = [
0.600698,
0.591385,
0.601863,
0.600698,
0.606519,
]
CellRank#
[78]:
F_hat_cellrank = pd.read_pickle(
"/home/mvinyard/github/scdiffeq-analyses/analyses/figure2/LARRY.fate_prediction/CellRank/ruitong_cellrank_dynamo/Seed0_termFate2.pickle"
)
adata.obs["combined_barcode"] = (
adata.obs["Library"].astype(str)
+ ":"
+ adata.obs["Cell barcode"].str.replace("-", "")
)
F_hat_cellrank_index = adata.obs.loc[
adata.obs["combined_barcode"].isin(F_hat_cellrank.index)
].index
F_hat_cellrank.index = F_hat_cellrank_index
F_hat_cellrank = F_hat_cellrank.div(F_hat_cellrank.sum(1), axis=0).fillna(0)
F_hat_cellrank = replace_undiff(F_hat_cellrank)
cellrank = compute_accuracy(F_obs, F_hat=F_hat_cellrank)
Dynamo#
[73]:
dynamo_csv_path = "/home/mvinyard/github/scdiffeq-analyses/analyses/figure2/LARRY.fate_prediction/dynamo/dynamo_fatebias.seed_0.csv"
adata.obs["combined_barcode"] = (
adata.obs["Library"].astype(str)
+ ":"
+ adata.obs["Cell barcode"].str.replace("-", "")
)
F_hat_dynamo = pd.read_csv(dynamo_csv_path, index_col=0)
F_hat_dynamo_index = adata.obs.loc[
adata.obs["combined_barcode"].isin(F_hat_dynamo.index)
].index
F_hat_dynamo.index = F_hat_dynamo_index
F_hat_dynamo = F_hat_dynamo.drop("confidence", axis=1)
F_hat_dynamo = F_hat_dynamo.drop("Undifferentiated", axis=1)
F_hat_dynamo = F_hat_dynamo.div(F_hat_dynamo.sum(1), axis=0).fillna(0)
F_hat_dynamo = replace_undiff(F_hat_dynamo)
dynamo = compute_accuracy(F_obs, F_hat=F_hat_dynamo)
PRESCIENT#
[67]:
F_hat_prescient = pd.read_csv(
"/home/mvinyard/experiments/run_prescient/F_hat.csvs/F_hat.PRESCIENT.LARRY.all_data.all_kegg-softplus_2_400-1e-06.seed_1.train.best.pt.csv",
index_col=0,
)
kegg_indicator = {"yes": "KEGG+", "no": "KEGG-"}
PRESCIENT_results = {"KEGG+": [], "KEGG-": []}
paths = list(pathlib.Path("/home/mvinyard/data/").glob("prescient*.pickle"))
for path in paths:
F_hat_pr = _read_process_F_hat(path=path, pickle=True)
acc = larry.tasks.fate_prediction.metrics.multi_idx_accuracy(F_obs, F_hat=F_hat_pr)
test_acc = acc.loc["unique_test.all_fates"].iloc[0]
# print(test_acc)
kegg_key = kegg_indicator[path.name.split("KEGG")[-1].split(".pickle")[0]]
PRESCIENT_results[kegg_key].append(test_acc)
PRESCIENT = pd.DataFrame(PRESCIENT_results)
scDiffEq#
[56]:
# scDiffEq - KEGG (no RVR)
f = pd.read_pickle(
"/home/mvinyard/github/scdiffeq-analyses/analyses/figure2/LARRY.fate_prediction/scDiffEq/scdiffeq_simout.pickle"
)
scDiffEq_KEGGNeg = {}
for seed in range(5):
key = ("scDiffEq1", "mu2000-sigma800-ss1", f"seed{seed}", "KEGGno")
F_hat_raw = f[key]
F_hat_raw.index = F_obs.index
F_hat_filt = F_hat_raw.drop("Undifferentiated", axis=1)
F_hat_norm = F_hat_filt.div(F_hat_raw.sum(1), axis=0).fillna(0)
F_hat_sdq = replace_undiff(F_hat_norm)
acc = larry.tasks.fate_prediction.metrics.multi_idx_accuracy(F_obs, F_hat=F_hat_sdq)
scDiffEq_KEGGNeg[seed] = acc.loc["unique_test.all_fates"].iloc[0]
# scDiffEq + KEGG (no RVR)
sdq_kegg = [
0.6239813736903376,
0.4947613504074505,
0.5273573923166472,
0.4947613504074505,
0.5064027939464494,
]
# scDiffEq + KEGG + RVR=2.5
project_path = pathlib.Path(
"/home/mvinyard/experiments/fate_prediction.reg_velo/v2/LightningSDE-FixedPotential-RegularizedVelocityRatio"
)
project = sdq.io.Project(project_path)
versions = [
getattr(project, attr) for attr in project.__dir__() if attr.startswith("version_")
]
best = {}
for version in versions:
version_accuracy = sdq_an.parsers.VersionAccuracy(version)
Vr = version.hparams["velocity_ratio_params"]["target"]
if Vr == 2.5:
best[version._NAME] = version_accuracy.best_test_from_train[["train", "test"]]
sdq_Vr2pt5 = pd.DataFrame(best).T["test"].values
org_results = get_organized_results(project_path)
best_results = get_best_results(org_results)
scDiffEq = {
"-KEGG": list(scDiffEq_KEGGNeg.values()),
"+KEGG": sdq_kegg,
"+KEGG+RVR2pt5": list(sdq_Vr2pt5),
}
torch-PBA#
[23]:
TorchPBA = {}
for path in glob.glob(
"/home/mvinyard/github/scdiffeq-analyses/analyses/figure2/LARRY.fate_prediction/PBA/PBA.F_hat_unfilt*.csv"
):
path_ = pathlib.Path(path)
seed = int(path_.name.split("seed_")[-1].split(".")[0])
F_hat = pd.read_csv(path_, index_col=0)
F_hat.index = F_hat.index.astype(str)
acc = larry.tasks.fate_prediction.metrics.multi_idx_accuracy(F_obs, F_hat=F_hat)
score = acc.loc["unique_test.all_fates"].iloc[0]
TorchPBA[seed] = score
Aggregate results#
[79]:
TorchPBA
[79]:
{5: 0.42607683352735737,
6: 0.5669383003492433,
0: 0.40279394644935973,
4: 0.3958090803259604,
3: 0.519208381839348,
1: 0.559953434225844}
[81]:
[81]:
[0.5087310826542492,
0.5389988358556461,
0.5389988358556461,
0.5133876600698487,
0.5261932479627474]
[115]:
results = {
"LR": LR,
"PBA": [0.363213],
"torch-pba": list(TorchPBA.values()),
"TIGON": [0.04084574723690534],
"CellRank": [cellrank],
"Dynamo": [dynamo],
"PRESCIENT": PRESCIENT_results["KEGG-"],
"PRESCIENT-KEGG": PRESCIENT_results["KEGG+"],
"scDiffEq": scDiffEq["-KEGG"],
"scDiffEq-KEGG": scDiffEq["+KEGG"],
"scDiffEq-KEGG-RV": scDiffEq["+KEGG+RVR2pt5"],
}
Plot#
[116]:
class StylishBoxPlot(ABCParse.ABCParse):
def __init__(
self,
colors: Optional[List[str]] = None,
widths: Optional[float] = None,
scatter_kw={
"alpha": 0.8,
"s": 25,
},
*args,
**kwargs
):
self.__parse__(locals())
@property
def colors(self):
if not hasattr(self, "_colors") or self._colors is None:
self._colors = list(cm.tab20.colors)
return self._colors
def _background_scatter(self, ax, data):
for en, (key, val) in enumerate(data.items()):
x = [key] * len(val)
if len(x) > 1:
x_vals = en + 1 + (np.random.random(len(x)) - 0.5) / 5
else:
x_vals = en + 1
ax.scatter(
x_vals,
val,
color=self.colors[en],
zorder=0,
ec="None",
rasterized=False,
**self._scatter_kw,
)
def _background_boxplot(self, ax, data):
x = list(data.keys())
y = list(data.values())
x = np.arange(len(y)) + 1
bp = ax.boxplot(
y,
positions=x,
patch_artist=True,
showmeans=True,
showfliers=False,
meanline=True,
zorder=1,
widths=self._widths,
)
for median in bp["medians"]:
median.set_visible(False)
for en, mean in enumerate(bp["means"]):
mean.set_c(self.colors[en])
for en, box in enumerate(bp["boxes"]):
box.set_facecolor(self.colors[en])
box.set_alpha(0.2)
for en, whisker in enumerate(bp["whiskers"]):
whisker.set_c("None")
for en, cap in enumerate(bp["caps"]):
cap.set_c("None")
def _foreground_boxplot(self, ax, data):
y = list(data.values())
x = list(data.keys())
x = np.arange(len(y)) + 1
bp = ax.boxplot(
y,
positions=x,
patch_artist=True,
showmeans=False,
showfliers=False,
meanline=False,
zorder=2,
widths=self._widths,
)
for en, box in enumerate(bp["boxes"]):
box.set_facecolor("None")
box.set_edgecolor(self.colors[en])
colors_ = np.repeat(
np.array(self.colors), 2, axis=0
) # list(np.repeat(self.colors, 2))
for en, whisker in enumerate(bp["whiskers"]):
whisker.set_c(colors_[en])
for en, cap in enumerate(bp["caps"]):
cap.set_c(colors_[en])
for median in bp["medians"]:
median.set_visible(False)
def __call__(self, ax, data, *args, **kwargs):
self.__update__(locals())
try:
self._background_scatter(ax, data)
except:
print(data)
self._background_boxplot(ax, data)
self._foreground_boxplot(ax, data)
[117]:
colors = [
"#262626",
"#f9626c",
"#9d0610",
"#ffcc00",
"#99cc33",
"#669900",
"#f27f34",
"#eb5e28",
"#00b4d8",
"#0096c7",
"#0077b6",
]
[118]:
results = {key: ABCParse.as_list(val) for key, val in results.items()}
fig, axes = cp.plot(
height=0.5,
width=1,
title=["LARRY fate prediction benchmark accuracy"],
y_label=["Accuracy Score"],
x_label=["Method"],
)
ax = axes[0]
sbp = StylishBoxPlot(colors=colors, widths=0.65)
sbp(ax, results)
ax.set_ylim(0, 0.7)
ax.set_xticks(np.arange(1, 12))
xtl = ax.set_xticklabels(list(results.keys()), ha="right", rotation=45)
plt.savefig("LARRY.fate_prediction_benchmark_accuracy.svg", dpi=500)