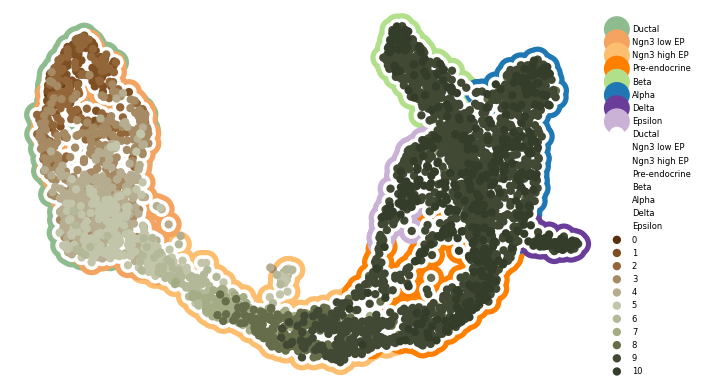
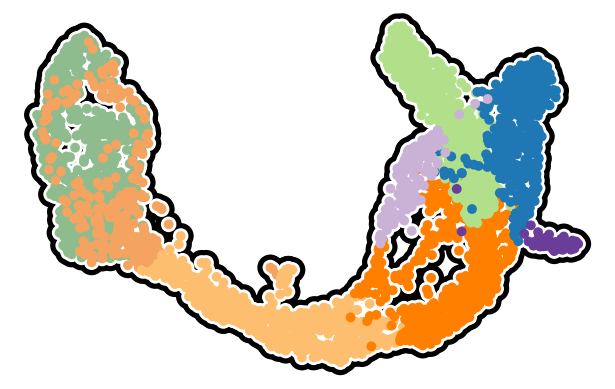
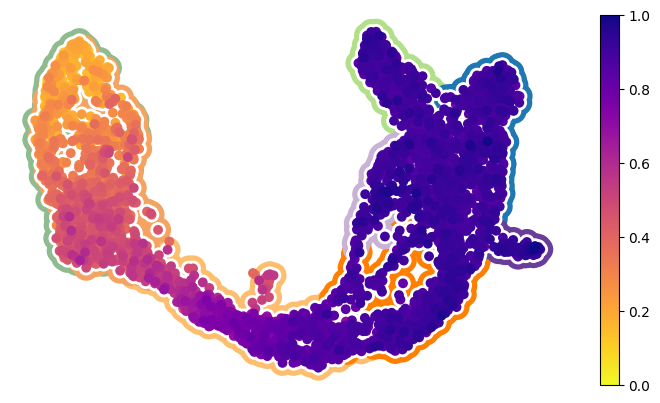
Figure S12ABC#
Plot pancreas data#
Import packages#
[1]:
%load_ext nb_black
import scdiffeq as sdq
import scdiffeq_analyses as sdq_an
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import cellplots as cp
import scanpy as sc
Load data#
[2]:
adata = sdq.datasets.pancreas()
/home/mvinyard/.anaconda3/envs/sdq-dev/lib/python3.9/site-packages/umap/umap_.py:1943: UserWarning: n_jobs value -1 overridden to 1 by setting random_state. Use no seed for parallelism.
warn(f"n_jobs value {self.n_jobs} overridden to 1 by setting random_state. Use no seed for parallelism.")
Prepare time#
cellrank has a handy implementation of CytoTRACE, which we’ll use to assign pseudotime.
[3]:
adata = sdq_an.tl.compute_CytoTRACE_pseudotime(adata)
Normalized count data: X, spliced, unspliced.
computing neighbors
finished (0:00:03) --> added
'distances' and 'connectivities', weighted adjacency matrices (adata.obsp)
computing moments based on connectivities
finished (0:00:07) --> added
'Ms' and 'Mu', moments of un/spliced abundances (adata.layers)
Next, we’ll bin the computed CytoTRACE pseudotime. We’ll use 10 non-t0 bins (i.e, 11 total)
[4]:
sdq_an.tl.bin_pseudotime(adata, pseudotime_key="ct_pseudotime", n_bins=10)
Plot data umaps#
[5]:
adata_pl = adata.copy()
adata_pl.obs["t"] = pd.Categorical(adata_pl.obs["t"])
clusters = adata.obs["clusters"].unique().sort_values()
clust_cmap = {i: j for i, j in zip(clusters, adata.uns["clusters_colors"])}
[6]:
fig, axes = cp.plot(1, 1, height=1, width=1.2, del_xy_ticks=[True], delete="all")
_ = cp.umap_manifold(
adata_pl,
ax=axes[0],
s_background=350,
s_cover=125,
)
zorder = {
list(clust_cmap.keys())[i]: 100 + i for i in range(len(list(clust_cmap.keys())))
}
_ = cp.umap(
adata_pl,
ax=axes[0],
groupby="clusters",
cmap=clust_cmap,
s=50,
force_zorder=zorder,
)
xu = adata_pl.obsm["X_umap"]
# img = axes[0].scatter(xu[:, 0], xu[:, 1], cmap=, zorder=101, ec="None", s=50)
# plt.colorbar(img)
plt.savefig("pancreas.cytotrace_manifold.svg")
[7]:
fig, axes = cp.plot(1, 1, height=1, width=1.4, del_xy_ticks=[True], delete="all")
_ = cp.umap_manifold(
adata_pl,
ax=axes[0],
groupby="clusters",
c_background=clust_cmap,
s_background=350,
s_cover=125,
)
xu = adata_pl.obsm["X_umap"]
c = adata.obs["ct_pseudotime"]
c_idx = np.argsort(c)
x, y = xu[c_idx, 0], xu[c_idx, 1]
img = axes[0].scatter(x, y, c=c[c_idx], cmap="plasma_r", zorder=101, ec="None", s=50)
plt.colorbar(img)
plt.savefig("pancreas.cytotrace_time.figure_ready.svg")
[8]:
t_colors = [
"#582f0e",
"#7f4f24",
"#936639",
"#a68a64",
"#b6ad90",
"#c2c5aa",
"#B3B998",
"#a4ac86",
"#656d4a",
"#414833",
"#333d29",
]
cmap = {i: j for i, j in zip(sorted(adata.obs["t"].unique()), t_colors)}
zorder = {i: 100 + i for i in range(len(t_colors))}
[9]:
fig, axes = cp.plot(1, 1, height=1, width=1.2, del_xy_ticks=[True], delete="all")
_ = cp.umap_manifold(
adata_pl,
ax=axes[0],
groupby="clusters",
c_background=clust_cmap,
s_background=350,
s_cover=125,
)
cp.umap(adata_pl, groupby="t", cmap=cmap, ax=axes[0], force_zorder=zorder)
plt.legend(facecolor="None", edgecolor="None", loc=(1, 0), markerscale=1, fontsize=6)
plt.savefig("pancreas.binned_cytotrace_time.figure_ready.svg")