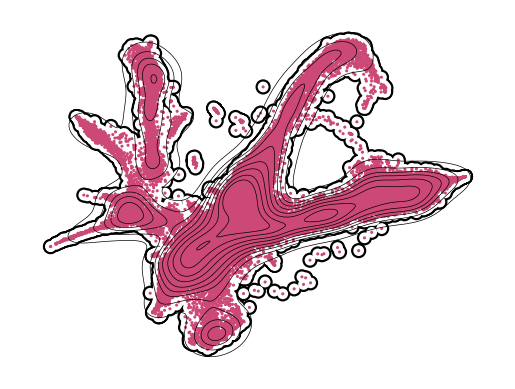
Figure S2c#
Import libraries#
[1]:
import anndata
import cellplots as cp
import larry
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import scdiffeq as sdq
import scdiffeq_analyses as sdq_an
import scipy.stats
import seaborn as sns
import sklearn
import torch
import umap
Read data#
This version of the LARRY dataset has not been split for test and train
[2]:
def download_in_vitro_adata():
in_vitro = larry.datasets.inVitroData()
adata = in_vitro.compose_adata()
return adata
[3]:
adata = download_in_vitro_adata()
- [ INFO ] | Added lineage x fate counts to: adata.uns['fate_counts']
- [ INFO ] | Added lineage-time occupance to: adata.uns['time_occupance']
- [ INFO ] | Fated cells annotated at: adata.obs['fate_observed']
- [ INFO ] | Fated cells (t=t0) annotated at: adata.obs['t0_fated']
- [ INFO ] | Added cell x fate counts to: adata.obsm['cell_fate_df']
[4]:
larry.datasets.klein_lab_pp_recipe.highly_variable_genes(adata)
[5]:
larry.datasets.klein_lab_pp_recipe.remove_cell_cycle_correlated_genes(adata)
[6]:
adata
[6]:
AnnData object with n_obs × n_vars = 130887 × 25289
obs: 'Library', 'Cell barcode', 'Time point', 'Starting population', 'Cell type annotation', 'Well', 'SPRING-x', 'SPRING-y', 'clone_idx', 'fate_observed', 't0_fated'
var: 'gene_ids', 'hv_genes', 'use_genes'
uns: 'fate_counts', 'time_occupance'
obsm: 'X_clone', 'cell_fate_df'
[7]:
adata = adata[:, adata.var['use_genes']].copy()
[8]:
adata.obs['train'] = adata.obs['Time point'].isin([2, 6])
adata.obs['test'] = adata.obs['Time point'].isin([4])
X_train = adata[adata.obs['train']].X
X_test = adata[adata.obs['test']].X
[9]:
ScalerModel = sklearn.preprocessing.StandardScaler()
PCAModel = sklearn.decomposition.PCA(n_components=50)
UMAPModel = umap.UMAP(n_components=2)
[10]:
X_train_scaled = ScalerModel.fit_transform(X_train.A)
X_test_scaled = ScalerModel.transform(X_test.A)
[11]:
X_train_pca = PCAModel.fit_transform(X_train_scaled)
X_test_pca = PCAModel.transform(X_test_scaled)
[12]:
X_train_umap = UMAPModel.fit_transform(X_train_pca)
X_test_umap = UMAPModel.transform(X_test_pca)
[13]:
adata_train = adata[adata.obs['train']].copy()
adata_test = adata[adata.obs['test']].copy()
adata_train.obsm['X_pca'] = X_train_pca
adata_train.obsm['X_umap'] = X_train_umap
adata_test.obsm['X_pca'] = X_test_pca
adata_test.obsm['X_umap'] = X_test_umap
adata_train.write_h5ad("adata_train.interpolation.larry.h5ad")
adata_test.write_h5ad("adata_test.interpolation.larry.h5ad")
[14]:
def _subset(adata_train, adata_test, t: int):
train_subset_idx = adata_train.obs.loc[adata_train.obs['Time point'] == t].index
test_subset_idx = adata_test.obs.loc[adata_test.obs['Time point'] == t].index
subset_train_X_pca = adata_train[train_subset_idx].obsm['X_pca']
subset_test_X_pca = adata_test[test_subset_idx].obsm['X_pca']
if t in [2, 6]:
return subset_train_X_pca
else:
return subset_test_X_pca
[15]:
Subsets = {}
for t in [2, 4, 6]:
Subsets[t] = _subset(adata_train, adata_test, t = t)
[16]:
sinkhorn = sdq.core.lightning_models.base.SinkhornDivergence()
[17]:
def compute_distance(X_pca_train, X_pca_test, N: int = 10, batch_size: int = 2048):
dist = {}
X_train = torch.Tensor(X_pca_train)
X_test = torch.Tensor(X_pca_test)
for i in range(N):
x_train_idx = np.random.choice(range(len(X_train)), batch_size)
x_test_idx = np.random.choice(range(len(X_test)), batch_size)
X_train_ = X_train[x_train_idx].to("cuda:0")
X_test_ = X_test[x_test_idx].to("cuda:0")
dist[i] = sinkhorn(X_train_, X_test_).detach().cpu().numpy()
return pd.Series(dist)
[18]:
Distances = {}
for ti, xi in Subsets.items():
Distances[ti] = {}
for tj, xj in Subsets.items():
Distances[ti][tj] = compute_distance(Subsets[ti], Subsets[tj], N = 25, batch_size = 2048)
print(ti, tj)
_df = pd.DataFrame(Distances[ti])
Distances[ti] = pd.DataFrame({"mean": _df.mean(), "std": _df.std()})
2 6
4 6
6 6
Distances from d2
[19]:
Distances[2]
[19]:
| mean | std | |
|---|---|---|
| 2 | 69.984912 | 24.334102 |
| 4 | 110.321152 | 16.750583 |
| 6 | 183.232344 | 17.898138 |
Distances from d4
[20]:
Distances[4]
[20]:
| mean | std | |
|---|---|---|
| 2 | 108.676318 | 12.697362 |
| 4 | 38.467209 | 6.045755 |
| 6 | 71.86564 | 6.566473 |
Distances from d6
[21]:
Distances[6]
[21]:
| mean | std | |
|---|---|---|
| 2 | 183.00168 | 18.961327 |
| 4 | 73.881343 | 9.951367 |
| 6 | 37.374524 | 7.5477 |
[22]:
time_cmap = sdq_an.pl.generate_temporal_cmap()
[23]:
t_coarse = time_cmap.colors[2:-2][::18]
t_coarse
[23]:
array([[0.979644, 0.854866, 0.142453],
[0.801855, 0.284626, 0.465971],
[0.227983, 0.016007, 0.604867]])
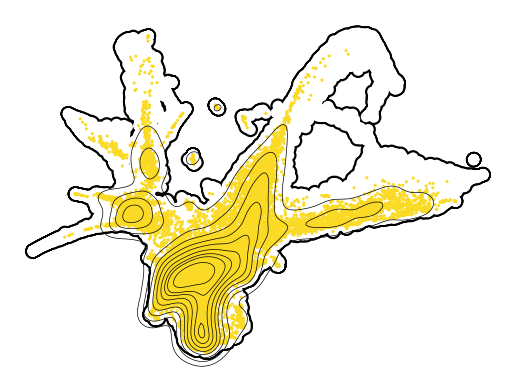
Plot d2, d6 (train)#
[24]:
for en, t in enumerate([2, 4, 6]):
if t != 4:
axes = cp.umap_manifold(adata_train)
cp.umap(adata_train[adata_train.obs['Time point'] == t], ax = axes[0], s = 5, c = t_coarse[en])
xu = adata_train[adata_train.obs['Time point'] == t].obsm['X_umap']
sns.kdeplot(x = xu[:,0], y = xu[:,1], c = "k", ax=axes[0], zorder = 301, linewidths=0.5)
plt.savefig(f"train.interpolation.timepoint_{t}.svg", dpi = 250)
plt.savefig(f"train.interpolation.timepoint_{t}.png")
plt.show()
/home/mvinyard/.anaconda3/envs/sdq/lib/python3.9/site-packages/cellplots/core/_umap.py:194: UserWarning: *c* argument looks like a single numeric RGB or RGBA sequence, which should be avoided as value-mapping will have precedence in case its length matches with *x* & *y*. Please use the *color* keyword-argument or provide a 2D array with a single row if you intend to specify the same RGB or RGBA value for all points.
ax.scatter(X_umap[:, 0], X_umap[:, 1], **KWARGS)
/home/mvinyard/.anaconda3/envs/sdq/lib/python3.9/site-packages/seaborn/distributions.py:1176: UserWarning: The following kwargs were not used by contour: 'c'
cset = contour_func(
/home/mvinyard/.anaconda3/envs/sdq/lib/python3.9/site-packages/cellplots/core/_umap.py:194: UserWarning: *c* argument looks like a single numeric RGB or RGBA sequence, which should be avoided as value-mapping will have precedence in case its length matches with *x* & *y*. Please use the *color* keyword-argument or provide a 2D array with a single row if you intend to specify the same RGB or RGBA value for all points.
ax.scatter(X_umap[:, 0], X_umap[:, 1], **KWARGS)
/home/mvinyard/.anaconda3/envs/sdq/lib/python3.9/site-packages/seaborn/distributions.py:1176: UserWarning: The following kwargs were not used by contour: 'c'
cset = contour_func(
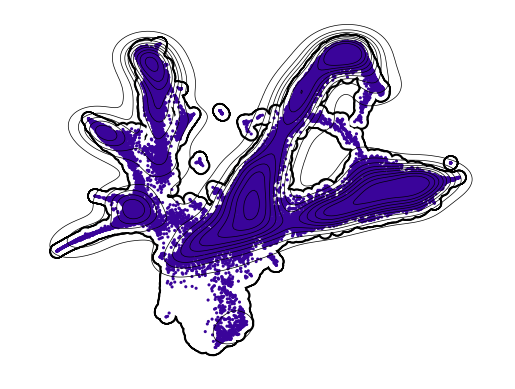
Plot d4 (test)#
[25]:
for en, t in enumerate([2, 4, 6]):
if t == 4:
axes = cp.umap_manifold(adata_test)
cp.umap(adata_test[adata_test.obs['Time point'] == t], ax = axes[0], s = 5, c = t_coarse[en])
xu = adata_test[adata_test.obs['Time point'] == t].obsm['X_umap']
sns.kdeplot(x = xu[:,0], y = xu[:,1], c = "k", ax=axes[0], zorder = 301, linewidths=0.5)
plt.savefig(f"test.interpolation.timepoint_{t}.svg", dpi = 250)
plt.savefig(f"test.interpolation.timepoint_{t}.png")
plt.show()
/home/mvinyard/.anaconda3/envs/sdq/lib/python3.9/site-packages/cellplots/core/_umap.py:194: UserWarning: *c* argument looks like a single numeric RGB or RGBA sequence, which should be avoided as value-mapping will have precedence in case its length matches with *x* & *y*. Please use the *color* keyword-argument or provide a 2D array with a single row if you intend to specify the same RGB or RGBA value for all points.
ax.scatter(X_umap[:, 0], X_umap[:, 1], **KWARGS)
/home/mvinyard/.anaconda3/envs/sdq/lib/python3.9/site-packages/seaborn/distributions.py:1176: UserWarning: The following kwargs were not used by contour: 'c'
cset = contour_func(