Figure 3H, I#
[1]:
%load_ext nb_black
import scdiffeq as sdq
import pathlib
import matplotlib.pyplot as plt
import cellplots as cp
import ABCParse
print(sdq.__version__, sdq.__path__)
0.1.2rc0 ['/home/mvinyard/github/scDiffEq/scdiffeq']
Load data, dimension reduction models#
[2]:
data_dir = pathlib.Path("/home/mvinyard/data/scdiffeq_data/human_hematopoiesis/")
h5ad_path = data_dir.joinpath("human_hematopoiesis.preprocessed.h5ad")
pca_model_path = data_dir.joinpath("human_hematopoiesis.pca_model.pkl")
umap_model_path = data_dir.joinpath("human_hematopoiesis.umap_model.pkl")
adata = sdq.io.read_h5ad(h5ad_path)
pca = sdq.io.read_pickle(pca_model_path)
umap = sdq.io.read_pickle(umap_model_path)
AnnData object with n_obs × n_vars = 1947 × 2477
obs: 'batch', '_time', 'nGenes', 'UMI', 't', 'n_genes', 'cell_type'
var: 'gene_name', 'n_cells', 'mean', 'std'
uns: 'log1p', 'h5ad_path'
obsm: 'X_pca', 'X_umap'
layers: 'X_l_TC', 'X_n_TC', 'X_orig', 'ambiguous', 'labeled_TC', 'sl_TC', 'sn_TC', 'spliced', 'total', 'ul_TC', 'un_TC', 'unlabeled_TC', 'unspliced'
Define some helper functions#
[3]:
class ManuscriptFigurePlot(ABCParse.ABCParse):
def __init__(self, cmap=None, color_quantile_cutoff: float = 0.01, *args, **kwargs):
self.__parse__(locals())
@property
def cmap(self):
if self._cmap is None:
self._cmap = {
"HSC": "#023047",
"MEP-like": "#fb8500",
"GMP-like": "#126782",
"Bas": "#219ebc",
"Mon": "#58b4d1",
"Neu": "#8ecae6",
"Meg": "#ffb703",
"Ery": "#fd9e02",
}
return self._cmap
@property
def scatter_kwargs(self):
return {
"s": 80,
"ec": "None",
"alpha": 0.4,
"vmin": self._adata.obs[self._c].quantile(self._color_quantile_cutoff),
"vmax": self._adata.obs[self._c].quantile(1 - self._color_quantile_cutoff),
}
def plot_manifold(
self,
adata,
groupby: str = "cell_type",
s_background=350,
s_cover=200,
*args,
**kwargs
):
fig, axes = cp.plot(
1, 1, height=1, width=1.2, del_xy_ticks=[True], delete="all"
)
axes = cp.umap_manifold(
adata,
groupby=groupby,
c_background=self.cmap,
s_background=s_background,
s_cover=s_cover,
ax=axes[0],
*args,
**kwargs,
)
self.axes = axes
self.axes[0].legend(
facecolor="None", edgecolor="None", markerscale=0.4, loc=(1.2, 0)
)
def plot_velocity(
self,
adata,
c="diffusion",
scatter_zorder=101,
stream_zorder=110,
return_axes=True,
png_dpi=500,
save=True,
svg_dpi=500,
scatter_kwargs={},
*args,
**kwargs
):
scatter_kw = self.scatter_kwargs.copy()
scatter_kw.update(scatter_kwargs)
self.axes[0] = sdq.pl.velocity_stream(
adata,
c=c,
ax=self.axes[0],
scatter_kwargs=scatter_kw,
scatter_zorder=scatter_zorder,
stream_zorder=stream_zorder,
return_axes=return_axes,
png_dpi=png_dpi,
save=save,
svg_dpi=svg_dpi,
*args,
**kwargs,
)
def __call__(
self,
adata,
groupby: str = "cell_type",
s_background=350,
s_cover=200,
c="diffusion",
scatter_zorder=101,
stream_zorder=110,
return_axes=True,
png_dpi=500,
save=True,
svg_dpi=500,
scatter_kwargs={},
*args,
**kwargs
):
self.__update__(locals())
self.plot_manifold(
adata=adata,
groupby=groupby,
s_background=s_background,
s_cover=s_cover,
)
self.plot_velocity(
adata=adata,
c=c,
scatter_zorder=scatter_zorder,
stream_zorder=stream_zorder,
return_axes=return_axes,
png_dpi=png_dpi,
save=save,
svg_dpi=svg_dpi,
scatter_kwargs=scatter_kwargs,
)
plt.show()
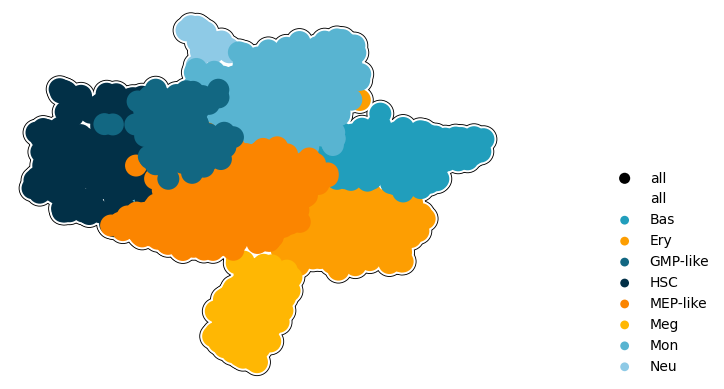
Figure 3H: plot the dataset UMAP manifold#
[4]:
cmap = {
"HSC": "#023047",
"MEP-like": "#fb8500",
"GMP-like": "#126782",
"Bas": "#219ebc",
"Mon": "#58b4d1",
"Neu": "#8ecae6",
"Meg": "#ffb703",
"Ery": "#fd9e02",
}
zorder = {
"HSC": 105,
"MEP-like": 106,
"GMP-like": 107,
"Bas": 104,
"Mon": 104,
"Neu": 103,
"Meg": 102,
"Ery": 101,
}
axes = cp.umap_manifold(
adata, c_background="k", s_background=400, c_cover="white", s_cover=350
)
axes = cp.umap(
adata, groupby="cell_type", cmap=cmap, s=250, ax=axes[0], force_zorder=zorder
)
axes[0].legend(facecolor="None", edgecolor="None", markerscale=0.4, loc=(1.2, 0))
plt.savefig("human_hematopoiesis.dynamo.svg", dpi=500)
Load project#
This “project” consists of five “versions”. In this case, each version is identical, wherein we run model training over five seeds (0 through 4).
[5]:
project = sdq.io.Project("./LightningSDE-FixedPotential-RegularizedVelocityRatio/")
Define helper functions#
[7]:
def aggr(df):
return df.filter(regex="sinkhorn").filter(regex="validation").dropna().sum(1).mean()
import pandas as pd
def aggr_total_loss(group_df, key="training"):
SD_loss = group_df.filter(regex="sinkhorn").filter(regex=key).dropna().sum(1).mean()
VR_loss = (
group_df.filter(regex="velo_ratio").filter(regex=key).dropna().sum(1).mean()
)
total_loss = VR_loss + SD_loss
return pd.Series({"total": total_loss, "VR": VR_loss, "SD": SD_loss})
def get_best_saved_ckpt(version):
grouped = version.metrics_df.groupby("epoch")
val = grouped.apply(aggr_total_loss)
saved_ckpts = list(version.ckpts.keys())
saved_ckpts.remove("last")
saved_ckpts.append(2499)
best_epoch = val["total"].loc[saved_ckpts].idxmin()
return best_epoch
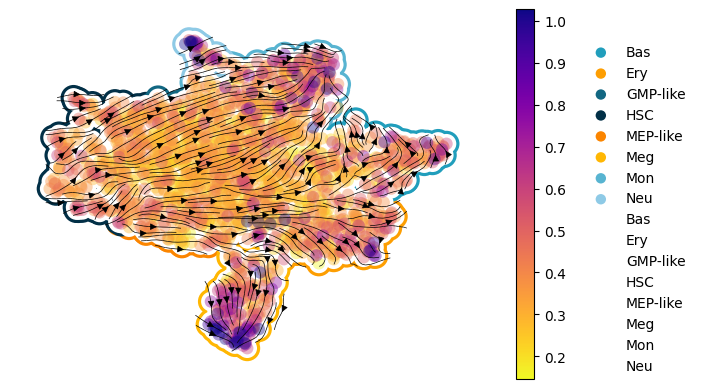
Figure 3I: scDiffEq velocity stream plots: human hematopoiesis#
[8]:
for v, version_path in project._VERSION_PATHS.items():
version = sdq.io.Version(version_path)
best_ckpt_epoch = get_best_saved_ckpt(version)
ckpt_path = version.ckpts[best_ckpt_epoch].path
model = sdq.io.load_model(adata=adata, ckpt_path=ckpt_path)
adata.uns["sdq_info"] = (
f"human_hematopoiesis.{v}.epoch_{best_ckpt_epoch}" # used for saving plot to a unique fname
)
model.drift(adata, silent=True)
model.diffusion(adata, silent=True)
sdq.tl.velocity_graph(adata, n_pcs=10)
plot = ManuscriptFigurePlot()
plot(adata)
Seed set to 0
- [INFO] | Input data configured.
- [INFO] | Bulding Annoy kNN Graph on adata.obsm['train']
- [INFO] | Using the specified parameters, LightningSDE-FixedPotential-RegularizedVelocityRatio has been called.
- [INFO] | Added: adata.obsp['distances']
- [INFO] | Added: adata.obsp['connectivities']
- [INFO] | Added: adata.uns['neighbors']
- [INFO] | Added: adata.obsp['velocity_graph']
- [INFO] | Added: adata.obsp['velocity_graph_neg']
- [INFO] | Saved to:
scdiffeq_figures/velocity_stream.human_hematopoiesis.version_1.epoch_1446.svg
scdiffeq_figures/velocity_stream.human_hematopoiesis.version_1.epoch_1446.png
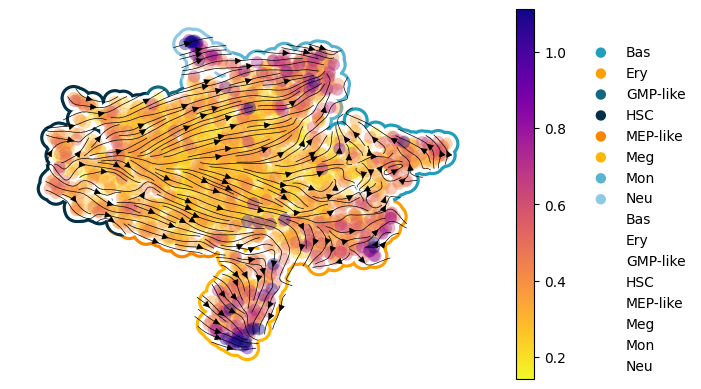
Seed set to 0
- [INFO] | Input data configured.
- [INFO] | Bulding Annoy kNN Graph on adata.obsm['train']
- [INFO] | Using the specified parameters, LightningSDE-FixedPotential-RegularizedVelocityRatio has been called.
- [INFO] | Updated: adata.obsp['velocity_graph']
- [INFO] | Updated: adata.obsp['velocity_graph_neg']
- [INFO] | Saved to:
scdiffeq_figures/velocity_stream.human_hematopoiesis.version_2.epoch_1464.svg
scdiffeq_figures/velocity_stream.human_hematopoiesis.version_2.epoch_1464.png
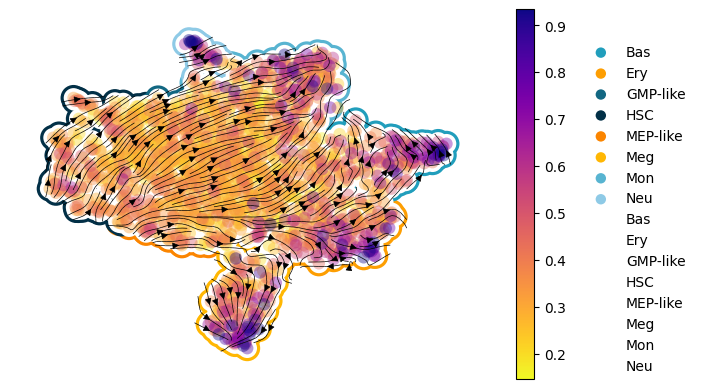
Seed set to 0
- [INFO] | Input data configured.
- [INFO] | Bulding Annoy kNN Graph on adata.obsm['train']
- [INFO] | Using the specified parameters, LightningSDE-FixedPotential-RegularizedVelocityRatio has been called.
- [INFO] | Updated: adata.obsp['velocity_graph']
- [INFO] | Updated: adata.obsp['velocity_graph_neg']
- [INFO] | Saved to:
scdiffeq_figures/velocity_stream.human_hematopoiesis.version_3.epoch_1543.svg
scdiffeq_figures/velocity_stream.human_hematopoiesis.version_3.epoch_1543.png
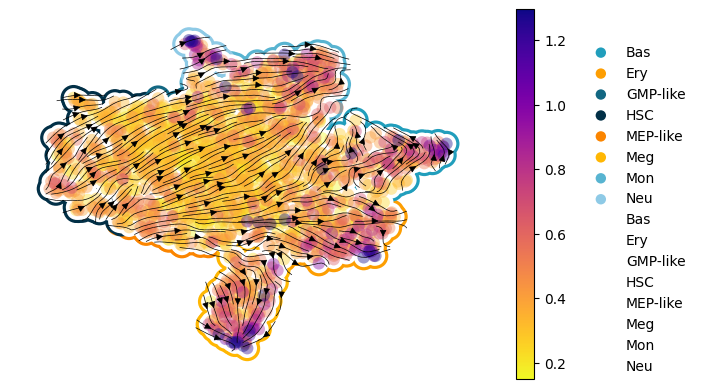
Seed set to 0
- [INFO] | Input data configured.
- [INFO] | Bulding Annoy kNN Graph on adata.obsm['train']
- [INFO] | Using the specified parameters, LightningSDE-FixedPotential-RegularizedVelocityRatio has been called.
- [INFO] | Updated: adata.obsp['velocity_graph']
- [INFO] | Updated: adata.obsp['velocity_graph_neg']
- [INFO] | Saved to:
scdiffeq_figures/velocity_stream.human_hematopoiesis.version_4.epoch_1951.svg
scdiffeq_figures/velocity_stream.human_hematopoiesis.version_4.epoch_1951.png
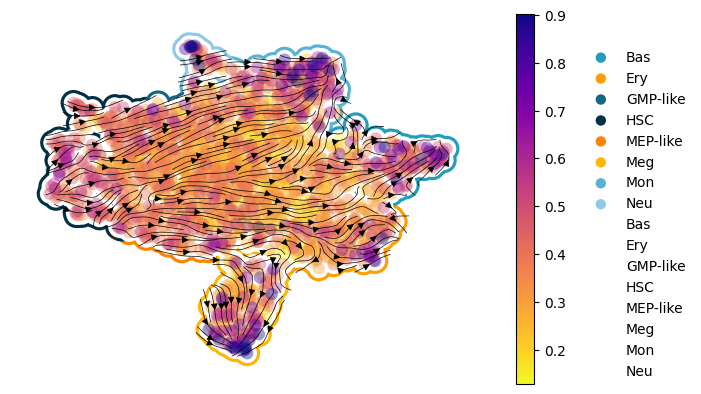
Seed set to 0
- [INFO] | Input data configured.
- [INFO] | Bulding Annoy kNN Graph on adata.obsm['train']
- [INFO] | Using the specified parameters, LightningSDE-FixedPotential-RegularizedVelocityRatio has been called.
- [INFO] | Updated: adata.obsp['velocity_graph']
- [INFO] | Updated: adata.obsp['velocity_graph_neg']
- [INFO] | Saved to:
scdiffeq_figures/velocity_stream.human_hematopoiesis.version_5.epoch_2264.svg
scdiffeq_figures/velocity_stream.human_hematopoiesis.version_5.epoch_2264.png