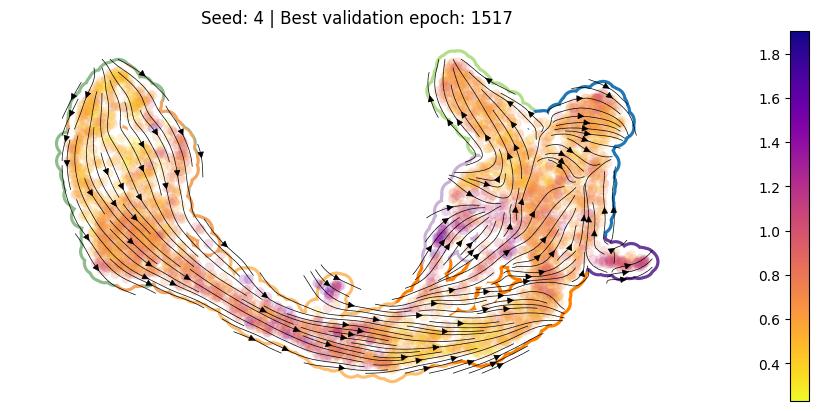
Figure 3M#
NOTE: This is the same notebook for FigureS12.
Import packages#
[1]:
import scdiffeq as sdq
import scdiffeq_analyses as sdq_an
import pandas as pd
import cellplots as cp
import matplotlib.pyplot as plt
Load project#
[2]:
project = sdq.io.Project("./LightningSDE-FixedPotential-RegularizedVelocityRatio/")
Organize loss values#
[3]:
SummarizedLoss = {}
for vname, version_path in project._VERSION_PATHS.items():
version = getattr(project, vname)
SummarizedLoss[vname] = sdq_an.parsers.SummarizedLoss(version=version)
Define function for the velocity_stream plot#
[4]:
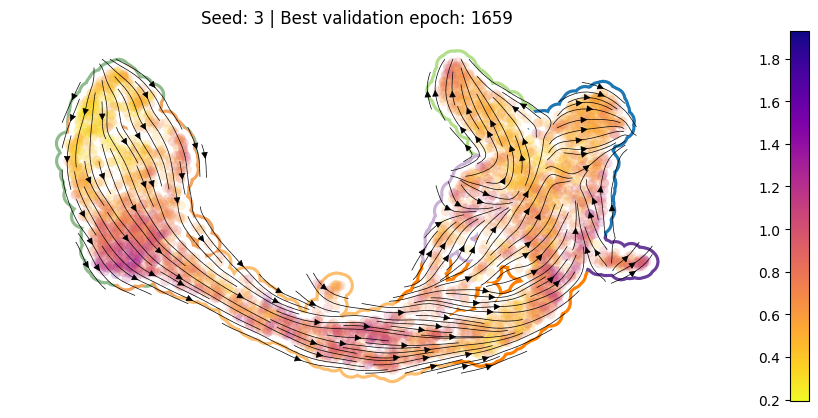
def pub_plot(adata, summarized_loss):
version = summarized_loss._version._NAME
v_int = int(version.split("_")[-1])
fig, axes = cp.plot(2, 2, width=1.4, del_xy_ticks=[True], delete="all")
_axes = cp.umap_manifold(
adata,
groupby="clusters",
c_background=clust_cmap,
s_background=350,
s_cover=200,
ax=axes[0],
)
best_epoch = summarized_loss.best_epoch
savename = f"seed_{v_int}.best_val_ckpt.epoch_{best_epoch}"
_ = sdq.pl.velocity_stream(
adata,
ax=axes[0],
c="diffusion", scatter_kwargs={"cmap": "plasma_r", "rasterized":True},
scatter_zorder=101,
stream_zorder=110,
return_axes=True,
cutoff_percentile=0.01,
save=False,
svg_dpi=500,
)
title = f"Seed: {v_int} | Best validation epoch: {best_epoch}"
axes[0].set_title(title)
img = axes[1].scatter(range(len(adata)), range(len(adata)), c = adata.obs['diffusion'], cmap = "plasma_r")
plt.colorbar(mappable = img, location="left")
plt.delaxes(axes[1])
plt.savefig(f"{savename}.svg", dpi=300)
plt.savefig(f"{savename}.png", dpi=300)
plt.show()
Load data#
[5]:
adata = sdq.io.read_h5ad("./data/Pancreas/endocrinogenesis_day15.h5ad")
AnnData object with n_obs × n_vars = 3696 × 27998
obs: 'clusters_coarse', 'clusters', 'S_score', 'G2M_score'
var: 'highly_variable_genes'
uns: 'clusters_coarse_colors', 'clusters_colors', 'day_colors', 'neighbors', 'pca', 'h5ad_path'
obsm: 'X_pca', 'X_umap'
layers: 'spliced', 'unspliced'
obsp: 'distances', 'connectivities'
Compute pseudotime, bin#
[6]:
adata = sdq_an.tl.compute_CytoTRACE_pseudotime(adata)
Normalized count data: X, spliced, unspliced.
computing neighbors
finished (0:00:03) --> added
'distances' and 'connectivities', weighted adjacency matrices (adata.obsp)
computing moments based on connectivities
finished (0:00:04) --> added
'Ms' and 'Mu', moments of un/spliced abundances (adata.layers)
[7]:
sdq_an.tl.bin_pseudotime(adata, pseudotime_key="ct_pseudotime", n_bins=10)
Prepare data for plotting#
[8]:
adata_pl = adata.copy()
adata_pl.obs["t"] = pd.Categorical(adata_pl.obs["t"])
clusters = adata.obs["clusters"].unique().sort_values()
clust_cmap = {i: j for i, j in zip(clusters, adata.uns["clusters_colors"])}
adata_pl.obs['t'] = adata_pl.obs['t'].cat.as_ordered()
Define a helper class for compatibility#
[9]:
class Model:
def __init__(self, ckpt_path):
self._ckpt_path = ckpt_path
@property
def DiffEq(self):
return sdq.io.load_diffeq(self._ckpt_path)
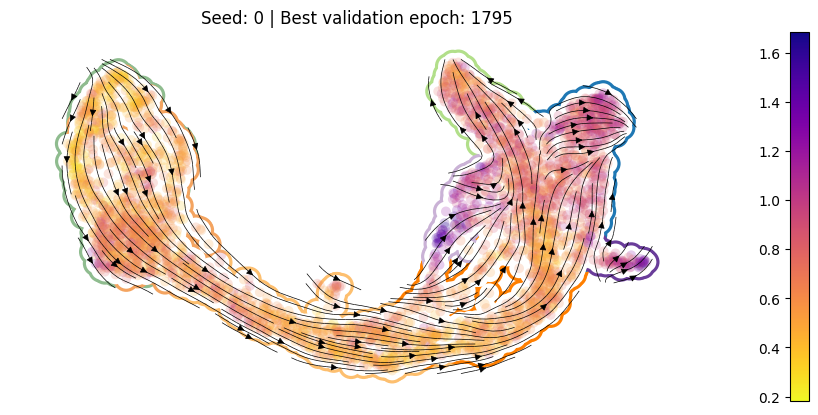
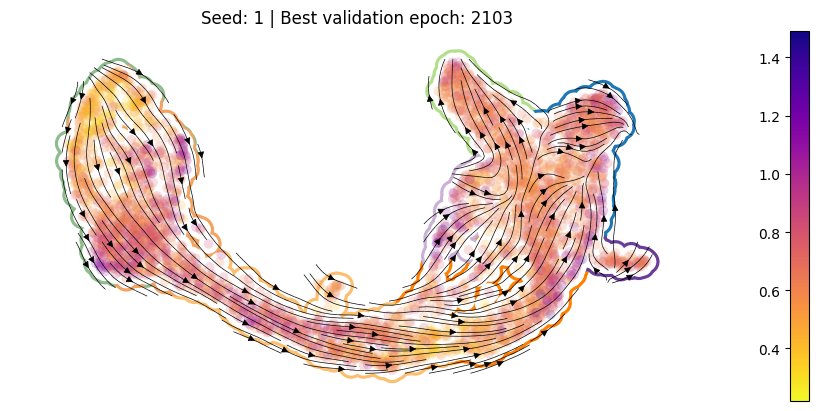
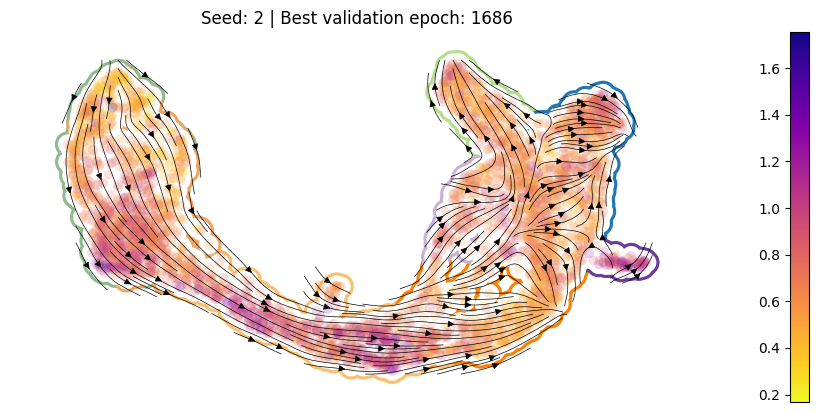
For each training run, plot the best ckpt#
Load the best validation
ckptCompute per-cell drift
Compute per-cell diffusion
Compute the velocity graph
Plot
[10]:
for en, (ver, summarized_loss) in enumerate(SummarizedLoss.items()):
ckpt_path = summarized_loss.best_ckpt.path
model = Model(ckpt_path = ckpt_path)
sdq.tl.drift(adata=adata_pl, model=model)
sdq.tl.diffusion(adata=adata_pl, model=model)
sdq.tl.velocity_graph(adata_pl)
pub_plot(adata_pl, summarized_loss)
- [INFO] | Added: adata.obsm['X_drift']
- [INFO] | Added: adata.obsm['drift']
- [INFO] | Added: adata.obsm['X_diffusion']
- [INFO] | Added: adata.obsm['diffusion']
- [INFO] | Added: adata.obsp['velocity_graph']
- [INFO] | Added: adata.obsp['velocity_graph_neg']
- [INFO] | Updated: adata.obsm['X_drift']
- [INFO] | Updated: adata.obsm['drift']
- [INFO] | Updated: adata.obsm['X_diffusion']
- [INFO] | Updated: adata.obsm['diffusion']
- [INFO] | Updated: adata.obsp['velocity_graph']
- [INFO] | Updated: adata.obsp['velocity_graph_neg']
- [INFO] | Updated: adata.obsm['X_drift']
- [INFO] | Updated: adata.obsm['drift']
- [INFO] | Updated: adata.obsm['X_diffusion']
- [INFO] | Updated: adata.obsm['diffusion']
- [INFO] | Updated: adata.obsp['velocity_graph']
- [INFO] | Updated: adata.obsp['velocity_graph_neg']
- [INFO] | Updated: adata.obsm['X_drift']
- [INFO] | Updated: adata.obsm['drift']
- [INFO] | Updated: adata.obsm['X_diffusion']
- [INFO] | Updated: adata.obsm['diffusion']
- [INFO] | Updated: adata.obsp['velocity_graph']
- [INFO] | Updated: adata.obsp['velocity_graph_neg']
- [INFO] | Updated: adata.obsm['X_drift']
- [INFO] | Updated: adata.obsm['drift']
- [INFO] | Updated: adata.obsm['X_diffusion']
- [INFO] | Updated: adata.obsm['diffusion']
- [INFO] | Updated: adata.obsp['velocity_graph']
- [INFO] | Updated: adata.obsp['velocity_graph_neg']