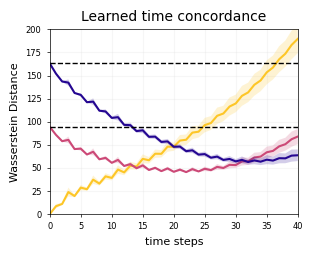
Figure 2F#
Simulated interpolation#
[ ]:
%load_ext nb_black
import matplotlib.pyplot as plt
import scdiffeq as sdq
import scdiffeq_analyses as sdq_an
import adata_query
import larry
import pandas as pd
import numpy as np
import tqdm.notebook
import ABCParse
import autodevice
import torch
import pathlib
import glob
from typing import Tuple
[1]:
h5ad_path = (
"/home/mvinyard/data/adata.reprocessed_19OCT2023.more_feature_inclusive.h5ad"
)
adata = sdq.io.read_h5ad(h5ad_path)
project_path = "/home/mvinyard/experiments/LARRY.full_dataset/LightningSDE-FixedPotential-RegularizedVelocityRatio"
project = sdq.io.Project(path=project_path)
InterpolationData = larry.tasks.interpolation.InterpolationData
sinkhorn = SinkhornDivergence()
SinkhornDivergence = sdq.core.lightning_models.base.SinkhornDivergence
class InterpolationTask(ABCParse.ABCParse):
def __init__(
self,
adata,
time_key="Time point",
use_key="X_pca",
t0=2,
n_samples=10_000,
lineage_key="clone_idx",
device=autodevice.AutoDevice(),
backend = "auto",
silent = False,
PCA = None,
*args,
**kwargs,
):
self.__parse__(locals())
self.data = InterpolationData(**self._DATA_KWARGS)
self.SinkhornDivergence = SinkhornDivergence(**self._SINKHORN_KWARGS)
@property
def _DATA_KWARGS(self):
return ABCParse.function_kwargs(
func=InterpolationData, kwargs=self._PARAMS
)
@property
def _SINKHORN_KWARGS(self):
return ABCParse.function_kwargs(
func=SinkhornDivergence, kwargs=self._PARAMS
)
def forward_without_grad(self, DiffEq):
"""Forward integrate over the model without gradients."""
with torch.no_grad():
X_hat = DiffEq.forward(self.data.X0, self.data.t)
return self._parse_forward_out(X_hat)
def forward_with_grad(self, DiffEq):
"""Forward integrate over the model retaining gradients."""
torch.set_grad_enabled(True)
X_hat = DiffEq.forward(self.data.X0, self.data.t)
return self._parse_forward_out(X_hat)
@property
def potential(self):
return "Potential" in str(self.DiffEq)
def _parse_forward_out(self, X_hat):
"""to account for KLDiv"""
if isinstance(X_hat, Tuple):
return X_hat[0]
return X_hat
def _dimension_reduce_pca(self, X_hat):
return torch.stack(
[torch.Tensor(self.PCA.transform(x)) for x in X_hat.detach().cpu().numpy()]
).to(self.device)
def __call__(self, trainer, DiffEq, *args, **kwargs):
self.__update__(locals())
if self.potential:
X_hat = self.forward_with_grad(DiffEq)
else:
X_hat = self.forward_without_grad(DiffEq)
if not self.PCA is None:
X_hat = self._dimension_reduce_pca(X_hat)
d4_loss = self.SinkhornDivergence(X_hat[1], self.data.X_test_d4).item()
d6_loss = self.SinkhornDivergence(X_hat[2], self.data.X_train_d6).item()
if not self.silent:
print(
"- Epoch: {:<5}| Day 4 loss: {:.2f} | Day 6 loss: {:.2f}".format(
DiffEq.current_epoch, d4_loss, d6_loss,
),
)
return d4_loss, d6_loss
AnnData object with n_obs × n_vars = 130887 × 2492
obs: 'Library', 'Cell barcode', 'Time point', 'Starting population', 'Cell type annotation', 'Well', 'SPRING-x', 'SPRING-y', 'clone_idx', 'fate_observed', 't0_fated', 'train'
var: 'gene_ids', 'hv_gene', 'must_include', 'exclude', 'use_genes'
uns: 'fate_counts', 'h5ad_path', 'time_occupance'
obsm: 'X_clone', 'X_pca', 'X_umap', 'cell_fate_df'
layers: 'X_scaled'
[5]:
class Results:
def __init__(self, Results):
self.d2 = pd.DataFrame([result["d2"] for i, result in Results.items()]).T
self.d4 = pd.DataFrame([result["d4"] for i, result in Results.items()]).T
self.d6 = pd.DataFrame([result["d6"] for i, result in Results.items()]).T
self.d2_d4 = pd.DataFrame([result["d2.d4"] for i, result in Results.items()]).T
self.d4_d6 = pd.DataFrame([result["d4.d6"] for i, result in Results.items()]).T
self.d2_d6 = pd.DataFrame([result["d2.d6"] for i, result in Results.items()]).T
self.d2_d2 = pd.DataFrame([result["d2.d2"] for i, result in Results.items()]).T
self.d4_d4 = pd.DataFrame([result["d4.d4"] for i, result in Results.items()]).T
self.d6_d6 = pd.DataFrame([result["d6.d6"] for i, result in Results.items()]).T
class PiecewiseDistance(ABCParse.ABCParse):
def __init__(
self,
adata,
n_samples: int = 10_000,
t=torch.linspace(2, 6, 41),
device: torch.device = autodevice.AutoDevice(),
*args,
**kwargs,
):
self.__parse__(locals())
self._df = self._adata.obs.copy()
self._clonal_df = self._df.loc[self._df["clone_idx"].notna()]
def sampling(self, group_df, N: int = 10_000):
if N > group_df.shape[0]:
replace = True
else:
replace = False
return group_df.sample(N, replace=True).index
def sample_indices(self, clonal_df):
print("Call clonal sampling")
return {
group: self.sampling(group_df)
for group, group_df in clonal_df.groupby("Time point")
}
@property
def X2(self):
if not hasattr(self, "_X2"):
self._X2 = adata_query.fetch(
self._adata[self.indices[2]],
key="X_pca",
torch=True,
device=self._device,
)
return self._X2
@property
def X4(self):
if not hasattr(self, "_X4"):
self._X4 = adata_query.fetch(
self._adata[self.indices[4]],
key="X_pca",
torch=True,
device=self._device,
)
return self._X4
@property
def X6(self):
if not hasattr(self, "_X6"):
self._X6 = adata_query.fetch(
self._adata[self.indices[6]],
key="X_pca",
torch=True,
device=self._device,
)
return self._X6
@property
def t(self):
return self._t.to(self._device)
def simulate(self, DiffEq):
return DiffEq.forward(self.X2, t=self.t)
@property
def divs(self):
if not hasattr(self, "_divs"):
divs = np.linspace(0, self._n_samples, 11).astype(int)
self._divs = (divs[:-1], divs[1:])
return self._divs
@property
def Z_hat(self):
if not hasattr(self, "_Z_hat"):
self._Z_hat = self.simulate(self._DiffEq)
return self._Z_hat
def forward(self, XA, XB, *args, **kwargs):
self.__update__(locals())
self._distances = []
div_i, div_j = self.divs
with torch.no_grad():
for i, j in zip(div_i, div_j):
self._distances.append(sinkhorn(XA[i:j], XB[i:j]))
return torch.stack(self._distances).detach().cpu().mean().item()
def compute_d2_distance(self):
return [self.forward(self.Z_hat[i], self.X2) for i in range(len(self.Z_hat))]
def compute_d4_distance(self):
return [self.forward(self.Z_hat[i], self.X4) for i in range(len(self.Z_hat))]
def compute_d6_distance(self):
return [self.forward(self.Z_hat[i], self.X6) for i in range(len(self.Z_hat))]
def compute_self_distances(self):
self.X2_X4 = self.forward(self.X2, self.X4)
self.X2_X6 = self.forward(self.X2, self.X6)
self.X4_X6 = self.forward(self.X4, self.X6)
self.X2_X2 = self.forward(self.X2, self.X2)
self.X4_X4 = self.forward(self.X4, self.X4)
self.X6_X6 = self.forward(self.X6, self.X6)
def compute_d2_d6(self):
return self.forward(self.X2, self.X6)
def compute_d4_d4(self):
return self.forward(self.X2, self.X4)
def compute_d4_d6(self):
return self.forward(self.X2, self.X6)
def __call__(self, DiffEq, N=5):
self.__update__(locals())
_Results = {}
for i in tqdm.notebook.tqdm(range(self._N)):
self.indices = self.sample_indices(self._clonal_df)
self.compute_self_distances()
_Results[i] = {
"d2": self.compute_d2_distance(),
"d4": self.compute_d4_distance(),
"d6": self.compute_d6_distance(),
"d2.d2": self.X2_X2,
"d4.d4": self.X4_X4,
"d6.d6": self.X6_X6,
"d2.d4": self.X2_X4,
"d4.d6": self.X4_X6,
"d2.d6": self.X2_X6,
}
del self._X2
del self._X4
del self._X6
del self._Z_hat
return Results(_Results)
[8]:
best_ckpts = sdq_an.parsers.summarize_best_checkpoints(project)
for vname, ckpt_path in best_ckpts["ckpt_path"].items():
save_path = pathlib.Path(f"sdq.simulate_interpolation.{vname}.pkl")
if not save_path.exists():
model = sdq.io.load_model(adata, ckpt_path=ckpt_path)
pw_distance = PiecewiseDistance(adata)
version_results = pw_distance(model.DiffEq)
sdq.io.write_pickle(version_results, save_path)
- [INFO] | Input data configured.
- [INFO] | Bulding Annoy kNN Graph on adata.obsm['train']
Seed set to 0
- [INFO] | Using the specified parameters, LightningSDE-FixedPotential-RegularizedVelocityRatio has been called.
Call clonal sampling
Call clonal sampling
Call clonal sampling
Call clonal sampling
Call clonal sampling
[11]:
d2 = pd.DataFrame(
[
sdq.io.read_pickle(path).d2.mean(1)
for path in glob.glob("sdq.simulate_interpolation.*.pkl")
]
).T
d4 = pd.DataFrame(
[
sdq.io.read_pickle(path).d4.mean(1)
for path in glob.glob("sdq.simulate_interpolation.*.pkl")
]
).T
d6 = pd.DataFrame(
[
sdq.io.read_pickle(path).d6.mean(1)
for path in glob.glob("sdq.simulate_interpolation.*.pkl")
]
).T
d2_d6 = pd.DataFrame(
[
sdq.io.read_pickle(path).d2_d6.mean(1)
for path in glob.glob("sdq.simulate_interpolation.*.pkl")
]
).mean()[0]
d2_d4 = pd.DataFrame(
[
sdq.io.read_pickle(path).d2_d4.mean(1)
for path in glob.glob("sdq.simulate_interpolation.*.pkl")
]
).mean()[0]
[12]:
import cellplots as cp
[13]:
time_cmap = sdq_an.pl.TimeColorMap()()
[14]:
len(time_cmap)
[14]:
41
[22]:
colors = [time_cmap[4], time_cmap[20], time_cmap[40]]
[35]:
fig, axes = cp.plot(
1,
1,
height=0.5,
width=0.5,
x_label=["time steps"],
y_label=["Wasserstein Distance"],
title=["Learned time concordance"],
)
ax = axes[0]
for en, d in enumerate([d2, d4, d6]):
mean = d.mean(1)
std = d.std(1)
lo = mean - std
hi = mean + std
ax.fill_between(
mean.index, lo, hi, color=colors[en], alpha=0.2, ec="None", zorder=2
)
ax.plot(mean, color=colors[en])
ax.hlines(d2_d6, 0, 40, lw=1, color="k", ls="--", zorder=3)
ax.hlines(d2_d4, 0, 40, lw=1, color="k", ls="--", zorder=3)
ax.grid(True, alpha=0.2, c="lightgrey")
ax.set_ylim(0, 200)
ax.set_xlim(0, 40)
ax.tick_params(axis="both", width=0.5, length=2.5)
_ = [spine.set_linewidth(0.5) for spine in list(ax.spines.values())]
plt.savefig("interpolation.stepwise_distance.svg", dpi=500)